For several years I have dreamt of ontologies and directed graph structures for representation of the logic in Soil Taxonomy. The express purpose of such work is that computer-aided taxonomy is “easier,” less prone to error, and enables more powerful reasoning about inherently abstract – but reasonably well-defined – concepts.
As an undergraduate in Dr. Schulthess’ soils classes at UCONN – the “graphy-ness” of Soil Taxonomy became apparent to me. I was perhaps “prepared” to see some of these patterns, but they were also revealed by careful explanation in class and in my work with the local NRCS MLRA office. I went on to cultivate that knowledge with soils coursework at UCDavis with Dr. Southard. Since then, I have found Soil Taxonomy useful as a scaffold for my learning about soils as a Soil Scientist. I feel I can now work to push our questions and the rigor we apply to a whole new level.
It is hard to conceive of what a system for traversing Soil Taxonomy logic will offer. As the taxonomy has grown more complex, there have been calls to simplify and justify various criteria. These efforts must be undertaken in some cases – but they should be informed by the best available analysis – one that leverages the logical structure of the Keys.
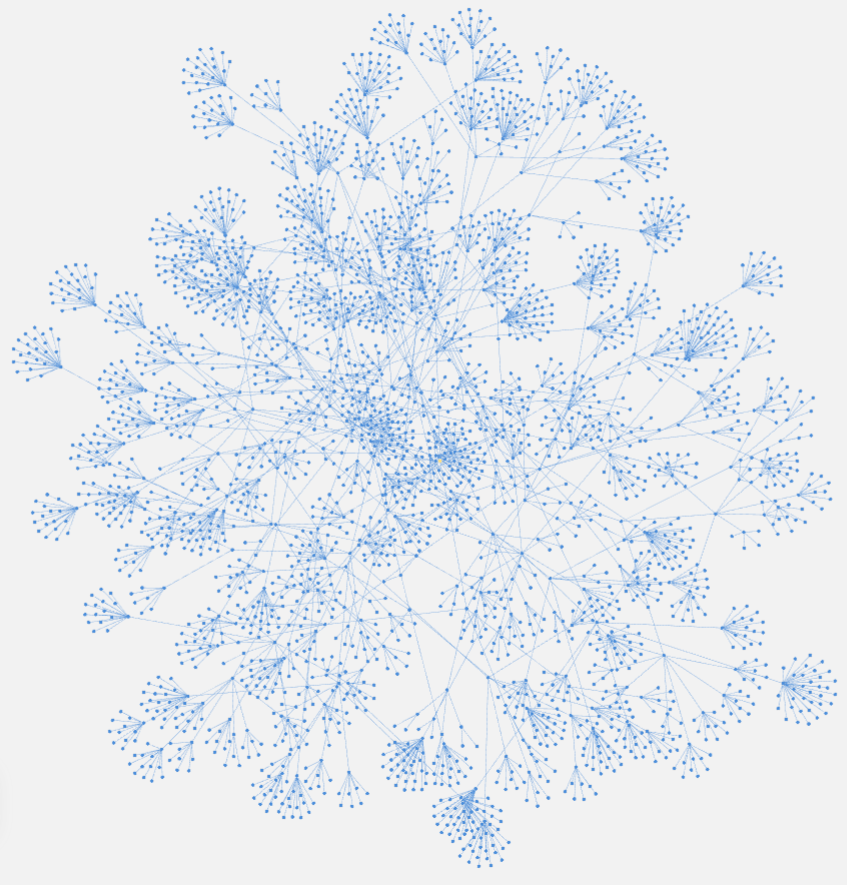
“Relationships are data, and data are beautiful.”
All 3082 subgroup-to-order level taxa in the 12th Edition Keys to Soil Taxonomy, with Soil Orders rooted to a common “Soil” node. Visualized with Neo4j Bloom.This blog post is one way of me communicating ideas that mostly live unspoken in my head. Also, it is an opportunity for “synergy” by fusing my recent work with existing technologies. Clearly, it is a continuation on the soil taxonomy topics that I love. I have been exploring further use of my REGEX parsed database of U.S. soil taxonomic information – gleaned from the official version of the Keys (12th edition). After addition of the Spanish language version of the “criteria” app, all of the kinks are worked out in the mapping of taxa and criteria to codes. For those who don’t know, I created this “database” a couple months ago for the Keys to Soil Taxonomy Shiny Apps. There are several prior blog posts about it, but here are the links to the apps for you to play with:
- KSTLookup - “criteria” app: http://brownag.shinyapps.io/KSTLookup
- KSTPreceding - “preceding taxa” app: http://brownag.shinyapps.io/KSTPreceding
These apps are alpha versions of “widgets” that expose the “parent-child” relationships implied by our hierarchical taxonomic system for soils in the U.S. The apps were intended to make people “think” about their changes to the fundamental structure and logic of the Keys.
For this new work, I will be using the Neo4j platform to begin more rigorously describing relationships between taxa. I will continue to work with taxa at the subgroup level or higher that form the backbone of taxonomy. Many of these principles will further apply to keys applied at lower (family/series/phase) levels. If you are interested in family and series concepts, the ncss-tech SoilTaxonomy repository will likely be of interest. Some of the R-based tools and ideas from there may be used in this work – depending on the capabilities of Neo4j-specific R tools – and it is likely that the static output from traversing the Neo4j database will resemble some of the data stored there.
Why Neo4j?
There are many reasons for choosing Neo4j out of the graph platforms currently available. It is an industry leader, free, plays nice with R, has a lovely IDE and visualization tools, it is wicked fast and scales to multiple cores, the cloud and more. If you want to read more about Neo4j check out some of these resources.
- Neo4j Graph Platform: https://neo4j.com/developer/graph-platform/
- Neo4j for R Developers and Data Scientists: https://neo4j.com/developer/r/#neo4j-r
Why Cypher?
The query language that I will be using is Cypher. Cypher is an open-source declarative language originally developed for Neo4j that has gained a foothold in the graph platform market. In fact it may be the best positioned for meeting the upcoming Graph Query Language Standard.
- Cypher Query Language: https://neo4j.com/developer/cypher/
- What is openCypher?: https://www.opencypher.org/
- What is in a Graph Query Language Standard?: https://www.gqlstandards.org/what-is-a-gql-standard
Trying Cypher out
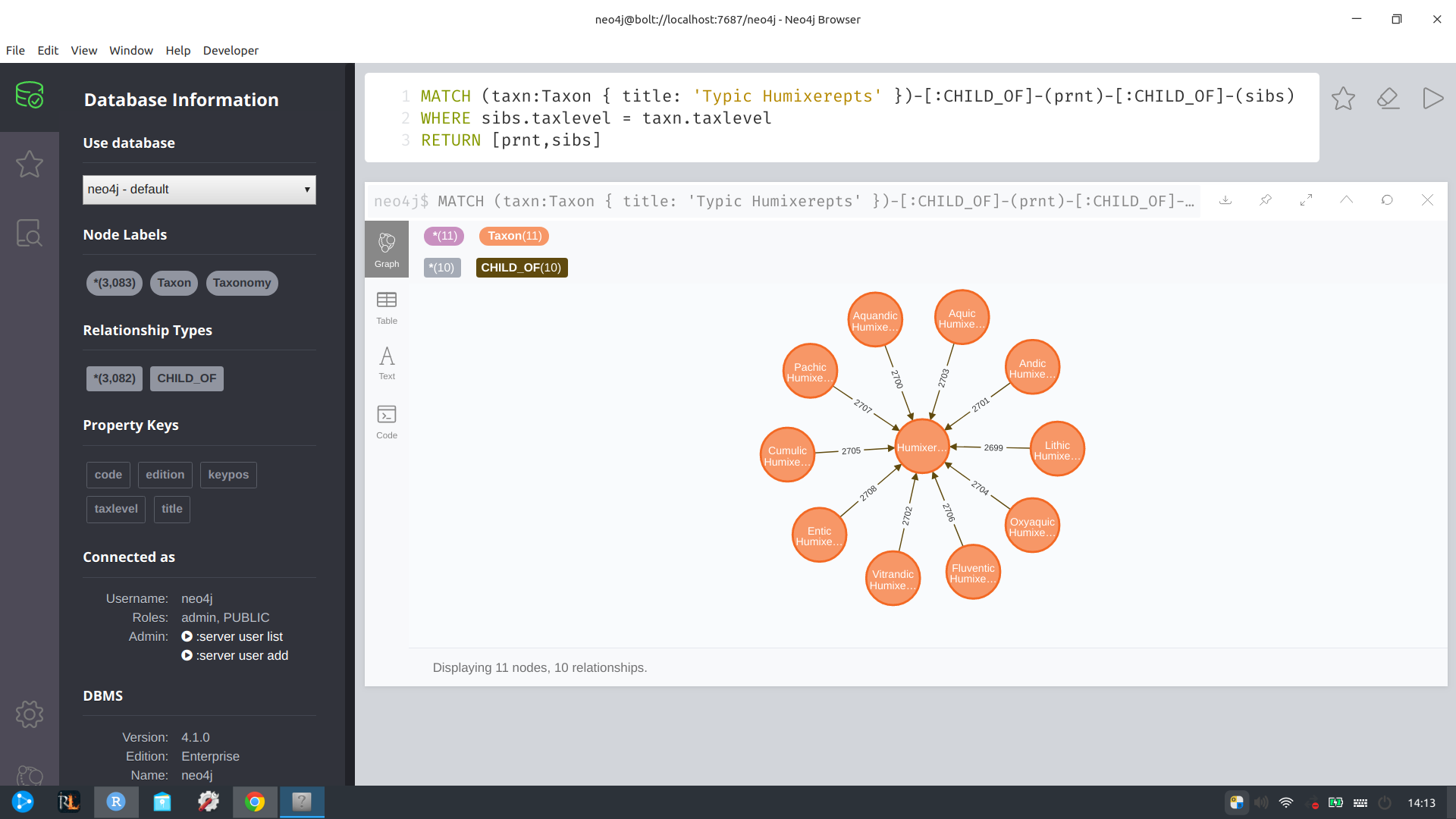
Here is a a sample Cypher query for taxa related to “Typic Humixerepts” that is run against my database for demonstration.
MATCH (taxn:Taxon { title: 'Typic Humixerepts' })-[:CHILD_OF]-(prnt)-[:CHILD_OF]-(sibs)
WHERE sibs.taxlevel = taxn.taxlevel
RETURN [prnt, sibs]I deliberately wrote this query to illustrate some features of Cypher. MATCH is similar to SQL SELECT, but we have the option to deal with both label- and property-level values. We create new objects whose relationships and properties we can reference throughout the rest of the query.
 The “directionality” (the key feature of Graph Databases) of relationships is expressed through syntax like R assignment operators. I wrote the connections in the above query in a direction-ambivalent way (
The “directionality” (the key feature of Graph Databases) of relationships is expressed through syntax like R assignment operators. I wrote the connections in the above query in a direction-ambivalent way (- v.s. <- or ->) to illustrate SQL-similarities with the option to use WHERE for constraints.
Rather than removing nodes via constraint on the taxlevel property, I can make use of the directional relationship between child and parent (prnt)<-[:CHILD_OF]-(sibs) to forgo the WHERE clause entirely.
To do this somewhat more Cypher-y, we just specify known directions of the CHILD_OF relationship.
MATCH (taxn:Taxon { title: 'Typic Humixerepts' })-[:CHILD_OF]->(prnt)<-[:CHILD_OF]-(sibs)
RETURN [prnt, sibs]This is most explicitly written with both relationships to parent specified (i.e. ->(prnt)<-). Since the example taxa (“Typic Humixerepts”) only have a single relationship ([:CHILD_OF]->(Humixerepts)) it does not matter, but the same could not be said for a higher-level taxa that have children and a parent. Or if our graph included taxa below subgroup.
A neat feature of Cypher is that you can concatenate results with the “Pythonic” (or Haskell-ic) syntax [x, y]. I use [prnt, sibs] to return both the parent (prnt) plus nodes that are the same taxonomic level “subgroup” (sibs) as the input (taxn).
Getting the data into a database
I wrote an R script generate a .cypher file that I can read in to create all of the data and relationships in my Neo4j database. There is nothing terribly novel about the script itself – just concatenating togther character strings. This sequence of Cypher commands is generated using some reworking of the constants and unique taxon codes used in the Keys to Soil Taxonomy.
The simplest way to do it is to set up relationships sequentially and individually. The order you create the nodes is relevant and interpreter will do some optimization of your input. I input ~3080 individual CREATE statements which are optimized to a single statement because of the relationships specified therein.
The first several auto-generated Cypher commands look like this:
CREATE (Soil:Taxonomy {title:'Soil Taxonomy', edition:12})
CREATE (Gelisols:Taxon {title:'Gelisols', code:'A', taxlevel:'order', keypos:1, parent:'Soil', parentcode:'*'})-[:CHILD_OF]->(Soil)
CREATE (Histels:Taxon {title:'Histels', code:'AA', taxlevel:'suborder', keypos:1, parent:'Gelisols', parentcode:'A'})-[:CHILD_OF]->(Gelisols)
CREATE (Turbels:Taxon {title:'Turbels', code:'AB', taxlevel:'suborder', keypos:2, parent:'Gelisols', parentcode:'A'})-[:CHILD_OF]->(Gelisols)
CREATE (Orthels:Taxon {title:'Orthels', code:'AC', taxlevel:'suborder', keypos:3, parent:'Gelisols', parentcode:'A'})-[:CHILD_OF]->(Gelisols)
...Visualizations
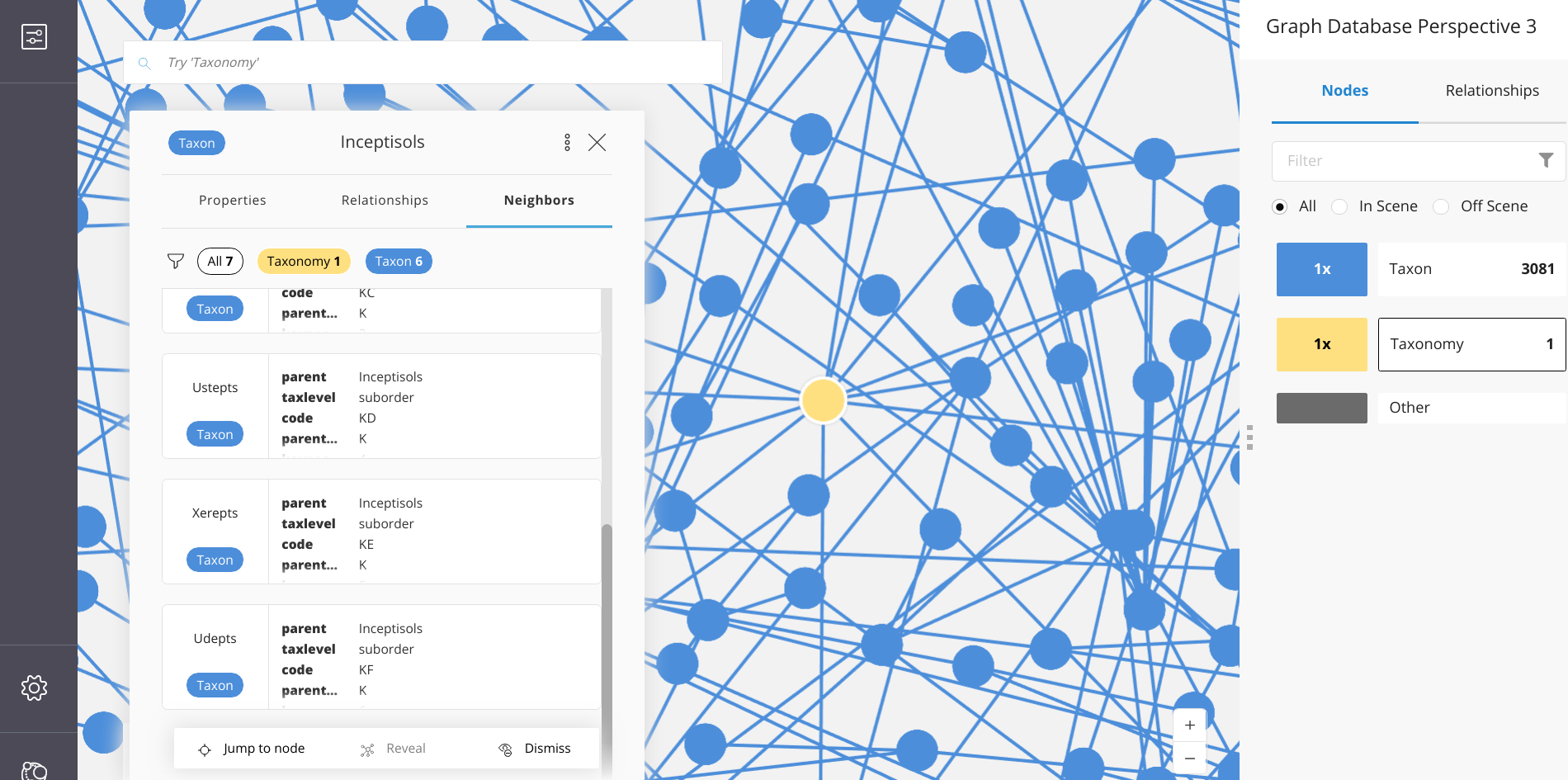
Using Neo4j Bloom, one can quickly make renderings of large networks – interact with nodes and relationships – and generate a variety of useful output.
Set up neo4j server on Ubuntu
Setting up a Neo4j server on Debian-based distributions is simple.
wget -O - http://debian.neo4j.org/neotechnology.gpg.key | apt-key add -
echo 'deb http://debian.neo4j.org/repo stable/' > /etc/apt/sources.list.d/neo4j.list
sudo apt update
sudo apt install neo4j
service neo4j status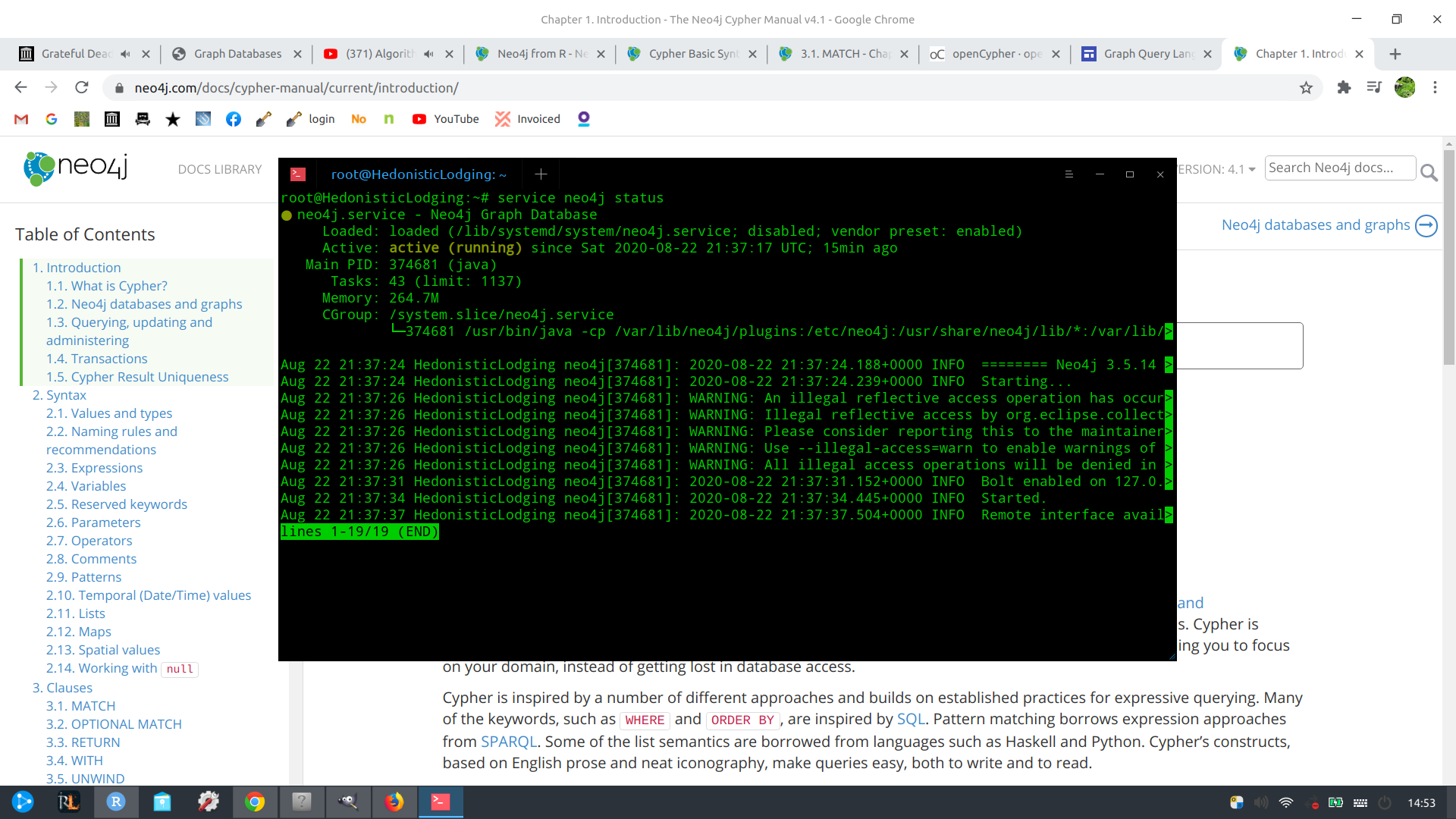
Future directions
I have developed a basic API leveraging the capabilities of Neo4j under-the-hood – this will be the topic of the next blog post.
I intend to dramatically expand semantic capabilities of this graph database using expanded REGEX parsing and new R scripts – currently there is only one relationship type – but once criteria are added there will be many new ways in which taxa begin to be “related.” Further, I will be implementing a variety of path-length type demonstrations that will be relevant to tracing logical paths to specific endpoints.
This platform opens up many possibilities for the Keys to Soil Taxonomy apps – which currently are using pre-computed taxonomic “trees.” I ultimately hope to be able to use these graph databases to “render” a variety of types of static output as JSON – either on the fly via Web APIs in response to unique constraints or pre-calculated for e.g. a Soil Taxonomy mobile app that does not rely on a persistent internet connection.
I’m going to spend a few days in the High Country to clear my head … hopefully briefly escape the heat and smoke we have here in CA – but then I will be following up with more blogs on this topic!
References, Source & Further Reading
Cypher refcard: https://neo4j.com/docs/cypher-refcard/current/
Cypher manual: https://neo4j.com/docs/cypher-manual/current/introduction/
Graph Databases – free eBook from O’Reilly: https://graphdatabases.com/
brownag/ncss-standards: https://github.com/brownag/ncss-standards
ncss-tech/SoilTaxonomy: https://github.com/ncss-tech/SoilTaxonomy
Keys to Soil Taxonomy - NRCS Soils: https://www.nrcs.usda.gov/wps/portal/nrcs/detail/soils/survey/class/?cid=nrcs142p2_053580
